With the REACT project, we wish to define host-pathogen interactions of three viral respiratory tract infections; influenza, respiratory syncytial virus (RSV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Objective
WP1 will ensure efficient and timely implementation of the project.
Description of work
In WP1, the project coordinator (PC) will ensure that project tasks, milestones and deliverables are achieved on time and in line with the allocated budget. The PC will furthermore ensure that periodic and final reports are submitted on time. The PC will be responsible for the communication between the consortium and the European Commission, and will oversee and management any potential conflict issues and mitigate them immediately and accordingly.
Objective
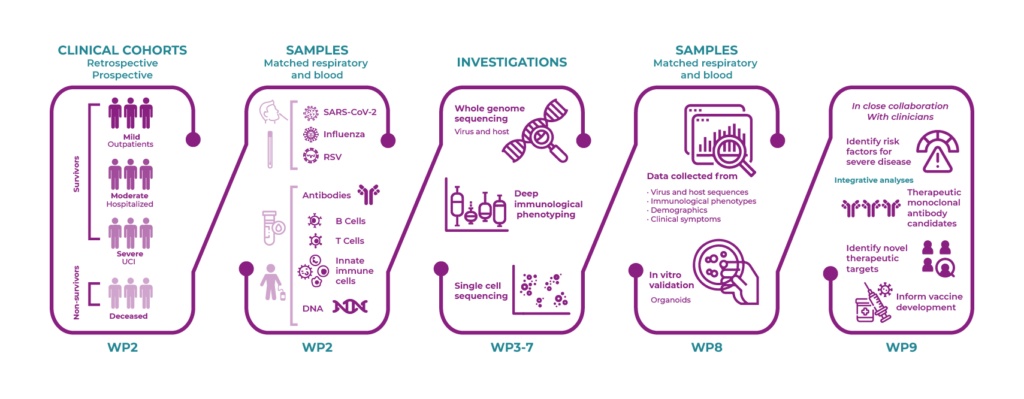
WP2 will collate and collect biological samples and data from research projects and national screening programs, to enable investigation of host-pathogen interaction and deep immunological characterization of emerging viral variants of concern.
Description of work
In WP2, we will coordinate the integration of existing cohorts, collections and biobanks, and the prospective collection of samples and live cells from virus-infected patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), influenza, or respiratory syncytial virus (RSV) in order to “live monitor” emerging viral variants and investigate host-pathogen interactions. Lastly, collaboration with existing EU-funded initiatives will be established.
Objective
WP3 will characterize and isolate the circulating variants of SARS-CoV-2, influenza, and RSV.
Description of work
In WP3, SARS-CoV-2, influenza and RSV will be characterized by whole genome sequencing (WGS) of samples from nasopharyngeal and oral swabs obtained through the Danish Sentinel Surveillance System and from samples provided through other available cohorts (WP2). Furthermore, deep bioinformatics analysis and variant calling will be performed, and selected virus variants will be isolated and cultured in BSL-3 facilities at SSI.
Objective
WP4 will determine the impact of host genetics for infection risk of different viral strains.
Description of work
In WP4, the genetic variants involved in disease susceptibility, disease severity and co-morbidities of the three viral strains will be identified, and also to what extent chronic disease and traits influences this. The association between infection and HLA genotype will be determined, and disease trajectory analysis will be performed.
Objective
WP5 will provide an in-depth understanding of T-cell dynamics to viral infections and disease protection.
Description of work
In WP5, the T cell derived immune components, both cellular and soluble, that confer disease protection will be identified. Furthermore, disease specific characteristics for all three viral strains will be determined, including identifying the T-cell receptor landscape to determine recognition of antigens and cross-reactivity to other viral antigens and its impact in viral infections.
Objective
WP6 will define serological and memory B-cell characteristics associated with mild versus severe disease following infection.
Description of work
In WP6, the overall contribution of virus-specific B-cell responses to beneficial versus pathological immune responses against SARS-CoV-2, influenza and RSV will be determined. Furthermore, the memory B-cell transcriptional programs associated with severe versus mild disease will be defined, and the beneficial and detrimental antibody characteristics identified.
Objective
WP7 will describe protective and deleterious innate immune events that predispose SARS-CoV-2 infected hospitalized patients to differential COVID-19 outcomes.
Description of work
In WP7, a cohort will be developed to store samples from black African COVID-19 patients in an HIV-1 endemic region in order to explore the clinical, radiological and laboratory parameters relating to initial presentation, clinical course and disease. Furthermore, the distinct humoral innate immune signatures associated with different clinical outcomes will be identified by transcriptomic profiling and targeted sequence analysis of innate immune genes.
Objective
WP8 will establish a preclinical platform based in patient-derived organoids for the validation of markers identified within the other WPs.
Description of work
In WP8, a human nasal epithelial (HNE) organoid culture system will be established as a validation platform and preclinical model. The HNE organoids will be infected with SARS-CoV-2, influenza and RSV, and single cell profiling characterization and immunofluorescence will be used to reveal unique and common signatures related to infection. The organoid models will enable investigation of host-pathogen interaction and deep immunological characterization of emerging viral variants of concern in the future by biobanking these samples within REACT biobanks.
Objective
WP9 will assess the clinical data and perform patient stratification and identification of prognostics biomarkers for personalized patient management.
Description of work
In WP9, for included cohorts (WP2), patient characteristics, treatment modalities and outcomes will be assessed. The diversity of host responses will be characterized at the level of genetic variants, molecular pathways and immunological interactions (WP5-8), and based on these data sources, a range of artificial-intelligence-based methods will be applied with the aim to find combinations of features for the respective outcomes when assessed individually. Lastly, the findings will be integrated in future clinical trials and practices.
Objective
WP10 will ensure dissemination, exploitation and communication of the results obtained in the REACT project.
Description of work
In WP10, dissemination of knowledge to the scientific community and to the society will be ensured, and specific knowledge communicated through the REACT webpage. Research results within REACT will be published in peer-reviewed scientific journals, and protocols and procedures shared to disseminate and fulfill best practice.
Objective
WP11 will ensure compliance with ethics requirements and guidelines through an established, independent Ethics Board.
Description of work
In WP11, an independent Ethics Board will be established to include members with relevant expertise to monitor the ethical concerns of the project. The ethics issues of the project primarily entail protection of vulnerable population, including children, and risk for stigmatization (HIV-positive patients) in research involving human participants, as well as protection of personal data and lack of evaluation of ethics risks. Periodic reports by the Ehtics Board will discuss how ethical issues were handled during a reporting period and assess possible newly emerged issues.
Objective
WP12 will use the lung-on-chip model to further characterise the host-pathogen interaction and response to viral infection.
Description of work
In WP12, an ex vivo platform based on human lung tissue will be established for a deeper mechanistic understanding of host-pathogen interaction, deep immunological characterization of emerging viral variants of concern and validation of therapeutic targets identified within REACT/REACT-HopOn.
